Coding Outreach Group Summer Workshop¶
Using Linux and Temple's Computing Cluster¶
07/22/21
Content Creator: Katie Jobson, Haroon Popal
Content Editor: Liz Beard
Description¶
This workshop will be an introduction to Temple University's Linux-based computing cluster, OwlsNest. Computing clusters are ideal for those who need to process large amounts of data, as those working with neuroimaging data often do.
We will use your newly aquired knowledge on Linux Machines and apply that to using OwlsNest.
Prerequisites¶
- Basic knowledge of bash scripting
- An OwlsNest account
To-do Before the Workshop¶
- Apply for an OwlsNest account with the High Performance Computing team here: https://www.hpc.temple.edu/
Workshop Objectives:¶
- Learn about Linux machines and their uses
- Gain the ability to log on and navigate OwlsNest
- Review how to setup a script to be submitted to the cluster
- Learn how to submit that script to the cluster and monitor jobs
Intro¶
from IPython.display import HTML
HTML('<iframe width="560" height="315" src="" frameborder="0" allowfullscreen></iframe>')
Linux Computers¶
I. What is a "linux"?¶
Linux is an operating system similar to UNIX, which Macs are built off of. The linux machines at Temple are usually a "high-end" computer which runs off the a linux operating system, such as Ubuntu. "High-end" refers to the computing resources of this machine which are very powerful compared to a standard desktop.
Example technical specifications¶
| Hardware | Details |
|---|---|
| CPU: | Intel(R) Xeon(R) Silver 4116 CPU @ 2.10GHz - 48 cores |
| Memory: | 256 GB |
| Hard Drive: | 10 TBs |
To give you a sense of what this means, a 2018 iMac's technical specifications are: CPU: 4.2 GHz Intel Core i7 - 8 cores Memory: 16 GB 2400 MHz DDR4 Hard Drive: 1 TB
In summary, the linux machine is not a normal computer. It is very powerful compared to a personal computer. As such, it should serve a different purpose for labs than a typical computer, and there is a "different way" to use it than a personal computer. With great power, comes great responsibility.
II. Why use a linux?¶
Different labs might have different motivations for using a linux machine. Below, I will discuss how our lab (the Olson lab) at Temple University uses our linux machine, and the purpose it serves.
The linux machine serves as a kind of server, both in terms of storing data and processing data. Users can "ssh" into the linux to access their data. They can complete preprocessing, analyses, anything else that can be done via the command line. However, it is recommended that the linux not be used for preprocessing, and instead, preprocessing should be done on Owlsnest. The reasoning behind this is that once you have a preprocessing pipeline locked down, it becomes mundane and does not need a lot of user intervention (you just a run few scripts, one after the other), nor does/should it change during the course of a project. Owlsnest is optimal for this because you would be able to schedule jobs to complete as others finish, without having to continually check the progress and completion of each step of your pipeline. Owlsnest also allows you to use large computing resources that are far greater than our linux, as one node from Owlsnest includes 28 cores, and Owlsnest has GPU resources. If Owslnest is used for preprocessing, this would free up our linux resources to allow users to run more "interactive" task with our more accessible linux machine. This would include analyses (first-level, second-level, stats, etc.) which often require a lot of development and tweaking over time. Users can use the linux machine to develop preprocessing scripts and pipelines and conducts tests. Owslnest can be tricky to beta test things, so the linux would be a good starting place as users can download relevant software and test them out. Once they get these working, they can transfer them to Owslnest for use.
III. How to use a linux¶
Logging in¶
You can log into the linux machine by using the following command:
ssh cla27562
ssh allows you to remote access a computer. The "cla27562" is the name of the linux machine. This is for when you are logging into a computer, where the username on that computer is your TU ID (i.e. you are using an Olson lab desktop). If you would like to log in from your personal laptop, you must be on the TU network (tusecurewireless), and use the following command:
ssh tu_id@cla27562.tu.temple.edu
If you wish to log into the linux from a computer that is not on the TU network, you must first remote access a computer that is on the network (i.e. you personal desktop in the lab). Please refer to Temple's remote access page for help with that.
Accessing the linux with the above commands will allow you to use the linux via the command line, but without graphics (can't use GUIs, open programs that have a graphic interface like matlab or fsleyes).
To log into the linux with graphics you can use the following command:
ssh -X cla27562
Before this command is functional, some things need to be set up on the user's personal computer (the computer you are logging in from). Information about that will be added... Also if the -X flag doesn't work, try -XY.
Copying files¶
To copy files from the linux, you can use the following command:
rsync -r cla27562:/file/to/be/copied /location/on/local/computer
The -r flag is needed to transfer directories (folders).
Regular files can be transferred without the flag.
Coding exercise 1: Copying files from linux¶
If you want to copy a file to the linux, reverse the two paths:
rsync ...
If you are copying files from Owlsnest to the linux, some special flags need to be run with rsync. For example:
rsync -rv --no-p --chmod=g+rx \
tuj96493@owlsnest.hpc.temple.edu:/home/tuj96493/work/fslprep \
/data/projects/11149BB/derivatives/
More about rsync here. Note that other commands can also be used, but I am most familiar with rsync.
Permissions¶
Our linux machine is set up with certain permissions that enforce the integrity of our system. Only members of the Olson lab have login access to the linux. Specifically, people have been added to the CLA Olson Lab group (via the lab manager). Users do not have admin (sudo) access to the linux. Since the linux will be used by multiple people, fail-safes need to be in place to guard against an increased likelihood of mistakes. Certain directories require sudo access to change, such as bids project directories. Installing software usually also requires sudo access. As such, if you need protected directories of files to be changed, or if you need software installed, speak to the linux administrator. Software should also be installed in a manner so that all users can use them, and this typically requires sudo access so that the software can be installed in a location all users have access to. Some softwares by default install in user specific directories (such as the user's home directory), but there are ways around this that are tricky, and so the administrator can help with this. Only users who have EXTENSIVE experience with linux-based operating systems and servers should request admin access.
Think Exercise 1:¶
What are some situations where you would want things installed for all users? What are some situations where you would want things installed for a user and not for everyone else?
Computing resources¶
As listed above, the linux machine has 48 cores (CPUs). These CPUs need to be monitored for usage. There are 256 GBs of memory, which is a lot, so memory usage should not be a problem. CPU usage can by monitored by using the "htop" command
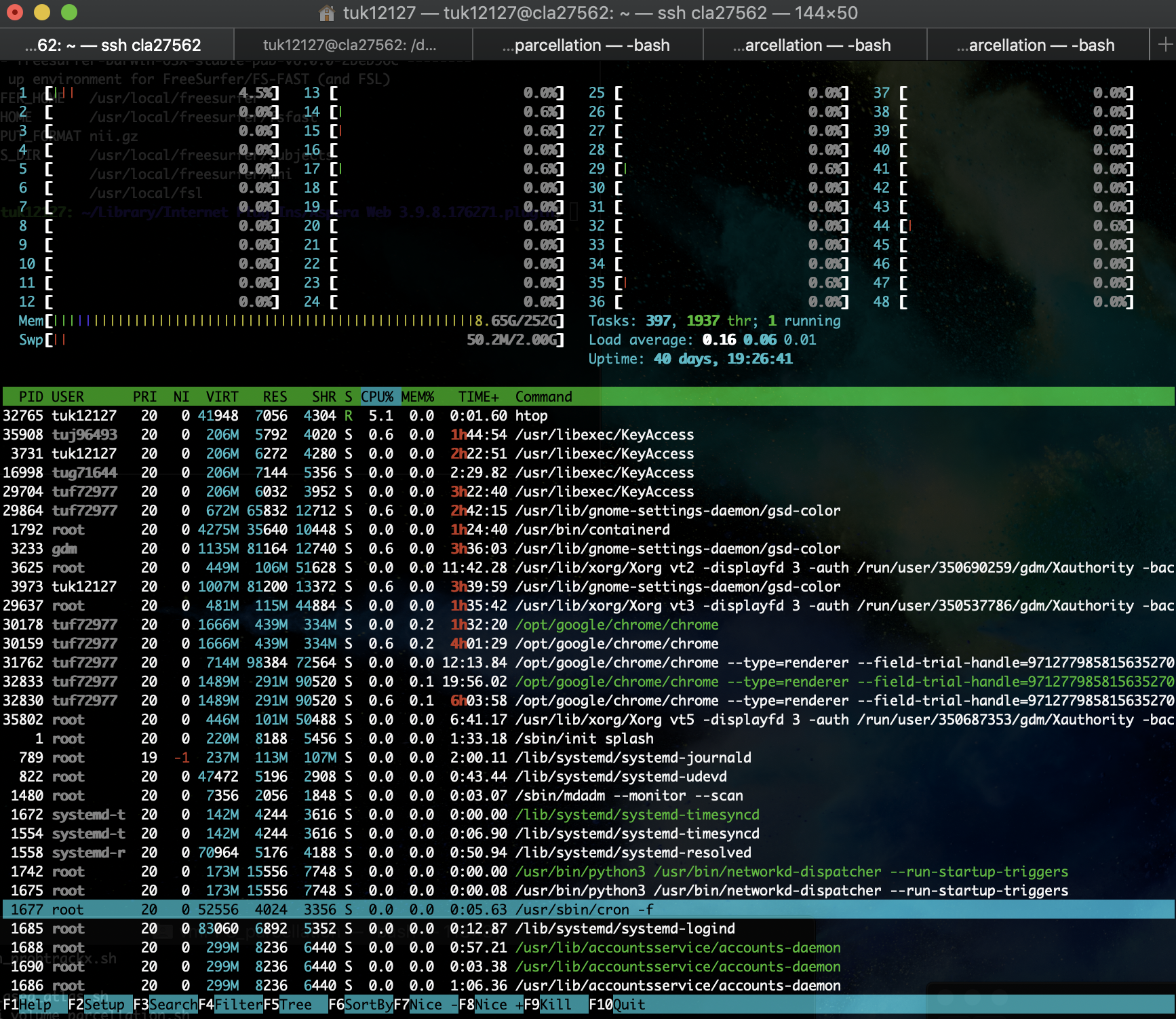
At the very top of this window, the 48 cores can be seen along with their usage. In this instance, only 1 CPU is being used at 4.5%. The memory bar seems like it is full, but the numbers to the right show that only 8.65GB out of 252GB (basically 256GB) are being used. In the second half of the window, the current processes that are running are being shown. Using this part is important to see which process is using what percentage of a CPU. For example, running this htop command is using 5.1% of one CPU. The USER column shows who is running that command (tuk12127, which is Haroon). This is helpful in knowing who to yell at for using too much resources (kidding). But it is helpful to know what resources are currently available and if you are able/should run whatever you are about to run. If all cores are being used, then you should wait until some free up. Generally a single command in one terminal window will use a single core. However, there are some process, especially in imaging analyses, that use multiple cores to speed things up. Be careful not to be a CPU hog, or to at least check with other users if you need to use a lot of resources. Generally, you should know the usage cost of whatever you are running on the linux. For example, when running FreeSurfer, I can configure it to run on four cores, and the command will run for 10 hours. This way I know when to expect my process to finish (in case someone needs the resources) and how to optimally use resources. Using more cores, for example, will not always give you the same benefit. See this example of how fmriprep uses resources:
Although you are not expect to create these plots for everything you plan on doing on the linux, you should have some kind of idea of how your processes run. This is done by testing your processes (i.e. run 1 subject, before running the other 49) and continually monitoring it. If you have any questions about how to manage resources for your work, talk to Haroon.
High Performance Computing - OwlsNest¶
Temple University offers a Linux-based computing cluster called OwlsNest. A local Linux machine is often expensive and needs knowledgable maintence. Having access to OwlsNest is the ideal situation for those who need high computing power but may not have the resources within their lab. Whether you have a large behavioral dataset or imaging data, this computing cluster can help you analyze your data without it taking weeks or months.
It is important to note that just because this all happens in a remote location, does not mean that there aren't real world consequences. Use OwlsNest with care, as these resources are used by the entire university. As such, every user needs to follow the same protocol and resource etiqeutte. With your personal computers, you are the only user. So you can set things up and run them however you want, because you are only effecting yourself. With Owlsnest, you would potentially be effecting hundreds of users across the entire university, and so strict protocols need to be followed. Detailed information can be found on the Owlsnest website.
Next, we will be showing you how to use your new Linux skills and apply them to navigating OwlsNest.
I. Logging In¶
Logging in to OwlsNest is fairly similar to the way we would log into a Linux machine via the command line. You will be using the 'ssh' command, along with your TUID. But there are some slight changes.
Logging on to a Linux machine can look like this:
ssh tuid1234@cla12345.tu.temple.edu
It takes this format because you are logging on to a computer on the Temple WiFi network. In this case, we will be logging on to a different type of network. To log on to the cluster, you will format it like this instead:
ssh tuid@owlsnest.hpc.temple.edu
Unlike logging on to a Linux machine, you do not have to be on the same WiFi network as the cluster. It can be accessed from anywhere!
Coding exercise 2: Log into your OwlsNest account¶
Now, try logging on for yourself!
Bonus question: What acknowledgement must you include if you publish with data you processed on OwlsNest?
ssh ...
II. Directory Structure on Owlsnest¶
While the cluster functions just like a Linux machine, there are certain limitations that are placed on users to ensure everyone gets enough storage space. There are different directories available, and those directories can hold a different amount of data and should be used for different things.
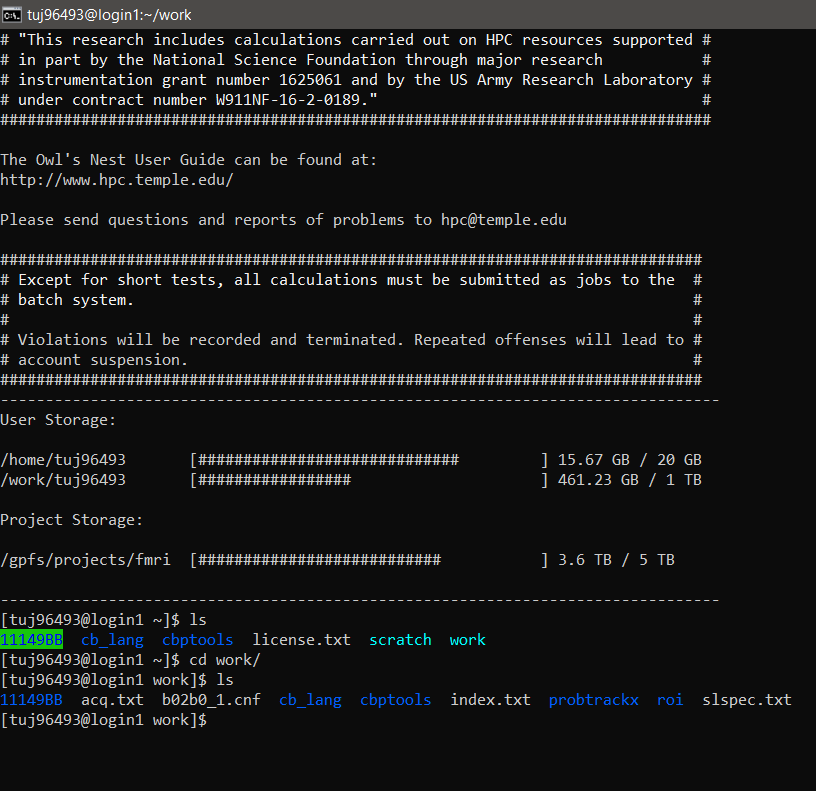
Everyone has three directories:
/home/tuj96493 - 20GB/home/tuj96493/work - TB/home/tuj96493/scratch - 500TB
If you have access to the folder that contains MRI projects, you also have:
/projects/fmri - 5TB
In order to keep to the size limits of each directory, it is recommended that you keep all scripts within your /home directory, and all of your data inputs and outputs in your /work directory. If you want to test some things out, and don't need the data forever, the /scratch folder is good for short-term things (note that the data is deleted from this folder after one month!).
It is also advised to have project directories to keep your account neat and easy to navigate. There is nothing worse than completing an analysis and coming back five months later, not knowing what folder contains your results!
Coding exercise 3: Create a study directory in /home and /work¶
Now, you can create a directory for yourself. Make one in your /home directory and one in your /work directory.
mkdir /home/tuid/studyname # replace tuid with your tuid
mkdir /home/tuid/work/studyname # replace tuid with your tuid
III. Setup a Script on OwlsNest¶
Next we are going to talk about how to write scripts to be run on the cluster. Because many people use OwlsNest, there is a queue system. So you must write a script that executes the commands you would like, and then you submit that script to the queue to be executed.
There are some lines of code that must be included at the top of your script to tell OwlsNest what to do with your script once it has been submitted. But first, let's create a script which we can edit:
cd /home/tuid/studyname
touch example.sh
Now that the script has been created, we can edit that script:
emacs example.sh
#!/bin/sh or #!/bin/python3 - /sh for bash scripting and /python3 for python shell
#PBS -n your_script_name - this is what your script will be called. Name it something meaningful so you know what is running at a glance.
#PBS -l walltime=1:00:00 - this is how much time you are requesting on the cluster. Take care when setting this - if your process has not finished running, the job will be killed by the system.
#PBS -q normal - this indicates what queue you are submitting to. To find more information about what resources each queue has, visit https://www.hpc.temple.edu/owlsnest2/batch_system/queues/
#PBS -l nodes=1:ppn=1 - this is where you request resources. It is best to only request one node per job, as it is unlikely that you have a job that will require more and you take resources from other individuals using the cluster
#PBS -n bae - this tells the system that you would like an email sent to you in case that your job runs (b), aborts (a), or when it ends (e). This is not required.
#PBS -M tuid@temple.edu - where to send emails if you set the previous command
#PBS -v subjects="5000" - any variable you would like to be exported to the job. I often put my subject list here
Make sure you save your work by typing:
Ctrl + x + c
One important thing to consider is the resources you request for your script. You need to be able to estimate how many cores that you will need, and about how much time it will take. You can do a few test runs to find out what you will need, but this is a skill that will come with use of the cluster.
To understand how to manage resources, it can be important to understand what those resources are. There are two hardware compenents of OwlsNest that we will talk about. One are nodes, and the other are cores. You can think of a node as a subsection of the cluster - a small computer within the cluster. A core can basically be thought of as a CPU. A normal desktop often only has one CPU. So, with multiple CPUs inside of it, a node is just a very powerful computer where your data can be processed.
One aspect of the resource management on OwlsNest that needs to be kept in mind is that when you request a node, no matter how many cores you intend on using, you will reserve that entire node. For instance; on the normal queue, one node contains 28 cores. You may denote that you would like to use "node=1:ppn=12", but you are still reserving all 28 cores. This is important, because if you cannot efficiently use the resources, you should be doing your anayses locally, or you should look into getting a Compute cluster account.
Next, we will talk about how to load different programs within the script so you may call on commands from those programs. To find a complete list of programs available on OwlsNest, type:
module avail
Bonus question: What version of one of your own frequently used programs is available as default on the cluster?
There are two ways to load a program. Either you can just list the program itself, and allow the cluster to call the default version of that program. Or you can tell it which version you would like to use. For example:
module load matlab
module load fsl/6.0.2
Now, executing commands on the cluster is slightly different depending on what you would like to achieve. Rather just writing out each command you would like to execute in your script, we create text files (specific to the scripts' job id on the cluster) that are then populated with those commands. This will allow each command to be executed one after another. Using these text files is a way to take advantage of the cluter's batch processing, which means that the script will execute multiple commands without input from the user. Here is an example of a script:
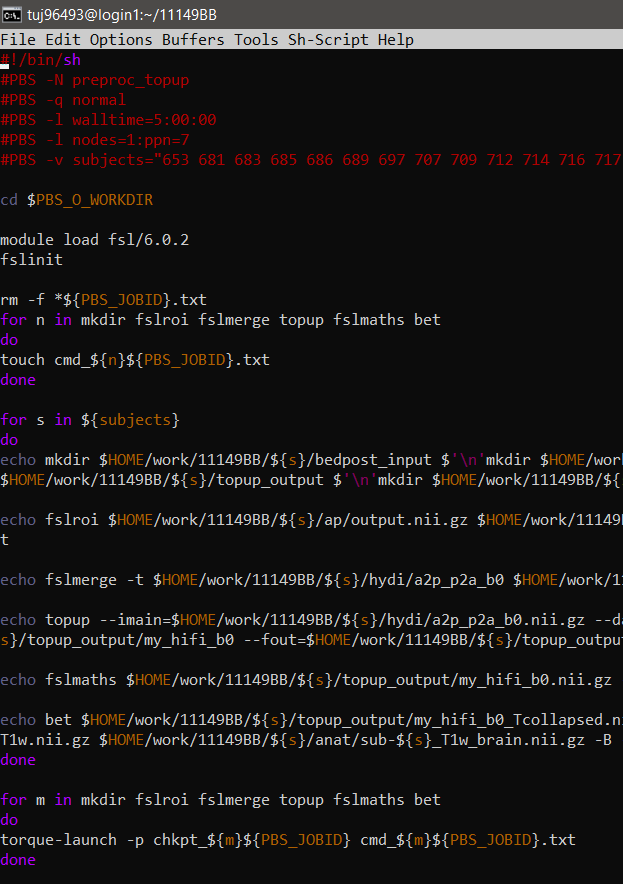
You can basically think of this as using a main script to execute other scripts. In this case, we are "creating" a script (aka our text files!) within our execution script, and then we can go on to execute them.
Here is a visual depiction of the batch processing system on OwlsNest:
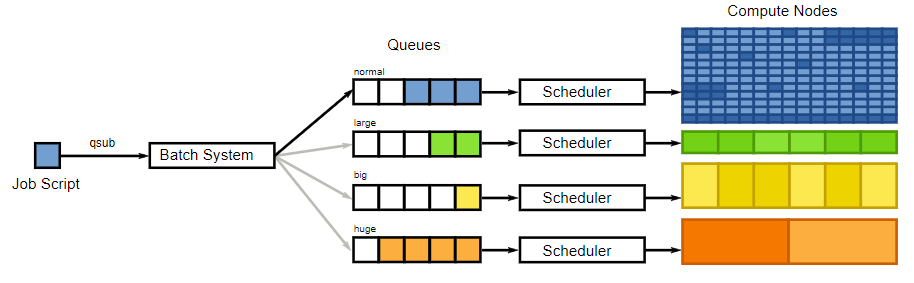
Alternatively, you can write an entire script (such as a python script) and execute it from within this script. By doing it this way, you are not longer able to take advantage of batch processing, so think wisely about how you would prefer to structure your scripts!
You cannot execute scripts directly from the command line, as you need to submit to the cluster's queue, which is why this execution script is a necessary step in using the cluster.
Coding exercise 4: Setup a script for an analysis you would run¶
Set up the script with the proper shell, give your script a name, and choose which queue you think would suit your processing needs the best!
IV. Submitting Scripts to the Cluster¶
Once you have completed you script, you are not able to submit it to the cluster! In order to submit it to the queue, you can execute this command:
qsub example.sh
Once it is submitted, there are a few ways to monitor its progress. One way is to log on to hpc.com and watch its progress from your account. It will tell you how much of the resources it is using, how long it has been running and how much time there is left.
If you want to check on your script in the command line, oyou can just use the command qstat. This will print out all of the jobs that are currently running on OwlsNest, their name, the user in charge of the script, whether it is queued, running or completed, and what queue it is in.
Troubleshooting is also often a necessary skill when working on the cluster.
Every script tht is submitted is assigned an ID (known as the ${PBS_JOBID}). To look at the error file for the job you have submitted, you can look at output named as such:
more your_script_name.e${PBS_JOBID}
This will print the contents of the error message. If it is empty, there was no error to be found. But, if there was any problem at any point in your script, it should be recorded here.
Think exercise 2: Requesting resources¶
Example problem where you need to run a script for 10 subjects. Two tasks need be done for each subject, and each task takes 10 hours per 8 cores. How many cores and time should you request?
Outro¶
from IPython.display import HTML
HTML('<iframe width="560" height="315" src="" frameborder="0" allowfullscreen></iframe>')
Supplementary¶
Suggested linux practices¶
Data Management¶
There are a few components to consider in regards to data storage for the lab. First, data needs to be stored in a central location so that all users who need access to a dataset, can access it with ease. Second, data needs to be backed up and archived to protect against mistakes and accidents. Lastly, because of these first two points, data will probably need to be moved around frequently.
Storage¶
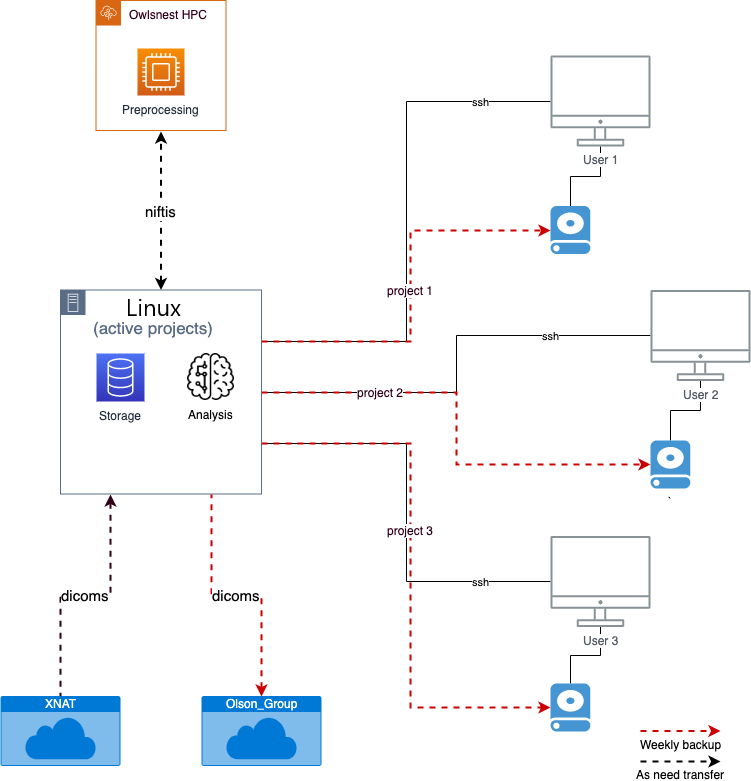
The linux machine can be used for storing "active project" data. This means projects that are in the data collection and analysis steps. Once a project as been completed (i.e. the paper has been published and no further analyses need to be run), the data should be removed from the linux (archiving data is covered below). With 10 TBs of storage, we will not be able to keep all data from the lab on the linux indefinitely. Project data should be kept in the "/data/projects/" directory on the linux as a single subdirectory. Projects with purely behavioral data (i.e. no imaging) can also be stored here, but behavioral data that was collected during an imaging project should be kept in the imaging project directory (e.g. bring-back study). Active project data can also be stored on user's personal computers, but that is up to the user. Portions of a dataset maybe be on Owlsnest for preprocessing. To move data between the linux, personal computers, and owslnest, the rsync command can be used. An example of this would be if you are scanning and need to preprocess your MRI data. You would first need to download the raw scans from XNAT onto your personal computer and rsync it to the linux. Then you would unpack the data and create the niftis. Then you would transfer the niftis to Owslnest, preprocess it, and then rsync it back to the linux for analysis. In this example, data might potentially be kept in two places, the linux and Owslnest. This redundancy adds some protection in case there are issues with the data at one of these locations. However, a more systematic backup solution is optimal.
Backups and Archiving¶
There are two ways in which data are systematically archived; raw dicoms are backed up onto the shared drive (Olson_Group), and project directories are backed up onto external hard drives. If possible, external hard drives will be connected to a single user's personal computer and entire project directories will be backed up via that personal computer to the external hard drive (red dashed lines in figure). If a personal computer is not available for this, hard drives can be linked connected to the linux machine to complete this back up. This is suboptimal as it can take up computing resources (although very minimal in practice), and having the backup hard drive directly connected to the linux machine can corrupt the external hard drive if the linux machine is corrected (this is a hypothetical issue and I may be paranoid). A single, exclusive, large (5TBs), external hard drive should be used for each project. Once a project is completed (no longer an "active project"), the external hard drive should be stored in a safe location for archiving (current the bookshelf in the back office).
Backups are completed using cronjobs. Cronjobs are commands that are scheduled to run at specific times. For each project on the linux machine, two cronjobs will be set to run rsync commands for the systematic backups. Users should have Haroon set up the backups for their projects.